Difference between revisions of "Computational Regulatory Genomics"
| (21 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<h1>Welcome to the Beer Lab!</h1> | <h1>Welcome to the Beer Lab!</h1> | ||
| − | [[File: | + | [[File:Beer_Michael_small.jpg]] [[File:EncodeNatureGraphic_small.png]] [[File:Beer_lab_plate_art_small.jpg]] |
| − | <h3>Research Interests: </h3> The ultimate goal of our research is to understand how genomic DNA sequence | + | <h3>Research Interests: </h3> The ultimate goal of our research is to understand how gene regulatory information is encoded in genomic DNA sequence. |
| − | We have recently made significant progress in understanding how DNA sequence features | + | We have recently made significant progress in understanding how DNA sequence features specify cell-type specific mammalian enhancer activity by using kmer-based SVM machine learning approaches. For details, see: |
* '''[http://www.horizonpress.com/genomeanalysis Mammalian Enhancer Prediction.]''' Lee D, Beer MA. 2014. Genome Analysis: Current Procedures and Applications. Horizon Press (in press) | * '''[http://www.horizonpress.com/genomeanalysis Mammalian Enhancer Prediction.]''' Lee D, Beer MA. 2014. Genome Analysis: Current Procedures and Applications. Horizon Press (in press) | ||
| Line 17: | Line 17: | ||
* '''[http://www.ncbi.nlm.nih.gov/pubmed/21875935 Discriminative prediction of mammalian enhancers from DNA sequence.]''' Lee D, Karchin R, and Beer MA. 2011. Genome Research 21:2167-2180. | * '''[http://www.ncbi.nlm.nih.gov/pubmed/21875935 Discriminative prediction of mammalian enhancers from DNA sequence.]''' Lee D, Karchin R, and Beer MA. 2011. Genome Research 21:2167-2180. | ||
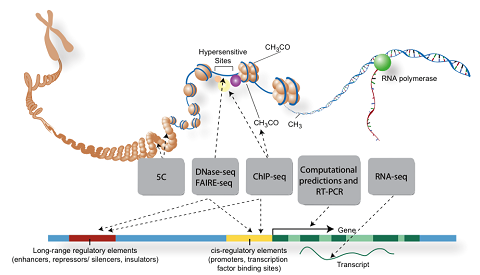
| − | + | Our work uses functional genomics DNase-seq, ChIP-seq, RNA-seq, and chromatin state data to computationally identify combinations of transcription factor binding sites which operate to define the activity of cell-type specific enhancers. We are currently focused on: | |
| − | * improving | + | * improving SVM methodology by including more general sequence features and constraints |
| − | * predicting the impact of SNPs on enhancer activity (delta-SVM) and GWAS | + | * predicting the impact of SNPs on enhancer activity (delta-SVM) and GWAS association for specific diseases |
| − | * experimentally | + | * experimentally assessing the predicted impact of regulatory element mutation in mammalian cells |
| − | * systematically determining regulatory | + | * systematically determining regulatory element logic from ENCODE human and mouse data |
| − | * using | + | * using this sequence based regulatory code to assess common modes of regulatory element evolution and variation |
| + | |||
| + | We are located in the McKusick-Nathans Institute for Genetic Medicine, and the Department of Biomedical Engineering, which has long been a leader in the development of rigorous quantitative modeling of biological systems, and is a natural home for graduate studies in Bioinformatics and Computational Biology at Johns Hopkins, including research in Genomics, Systems Biology, Machine Learning, and Network Modeling. | ||
<h3>[[Lab Members]]</h3> | <h3>[[Lab Members]]</h3> | ||
<h3>[[Publications]]</h3> | <h3>[[Publications]]</h3> | ||
| + | <h3>[[Postdoctoral Positions Available]]</h3> | ||
| + | <h3>About Computational Biology in JHU Biomedical Engineering:</h3> | ||
| + | The Department of Biomedical Engineering has long been a leader in the development of rigorous quantitative modeling of biological systems, and is a natural home for graduate studies in Bioinformatics and Computational Biology at Johns Hopkins. Students with backgrounds in Physics, Mathematics, Computer Science and Engineering are encouraged to apply. Opportunities for research include: Computational Medicine, Genomics, Systems Biology, Machine Learning, and Network Modeling. Graduate students in Johns Hopkins' Biomedical Engineering programs can select research advisors from throughout Johns Hopkins' Medical Institutions, Whiting School of Engineering, and Krieger School of Arts and Sciences. | ||
| + | |||
| + | <h3>[[Visit Some Computational Labs at Hopkins http://karchinlab.org/bme-compbio-jhu/]]</h3> | ||
| + | |||
| + | [[File:bmesmall.png]] | ||
Revision as of 01:44, 21 March 2014
Welcome to the Beer Lab!
Research Interests:
The ultimate goal of our research is to understand how gene regulatory information is encoded in genomic DNA sequence.
We have recently made significant progress in understanding how DNA sequence features specify cell-type specific mammalian enhancer activity by using kmer-based SVM machine learning approaches. For details, see:
- Mammalian Enhancer Prediction. Lee D, Beer MA. 2014. Genome Analysis: Current Procedures and Applications. Horizon Press (in press)
- Robust k-mer Frequency Estimation Using Gapped k-mers. Ghandi M, Mohammad-Noori M, and Beer MA. 2013. Journal of Mathematical Biology. (Epub ahead of print)
- kmer-SVM: a web server for identifying predictive regulatory sequence features in genomic datasets. Fletez-Brant C*, Lee D*, McCallion AS and Beer MA. 2013. Nucleic Acids Research 41: W544–W556.
- Integration of ChIP-seq and Machine Learning Reveals Enhancers and a Predictive Regulatory Sequence Vocabulary in Melanocytes. Gorkin DU, Lee D, Reed X, Fletez-Brant C, Blessling SL, Loftus SK, Beer MA, Pavan WJ, and McCallion AS. 2012. Genome Research 22:2290-2301.
- Discriminative prediction of mammalian enhancers from DNA sequence. Lee D, Karchin R, and Beer MA. 2011. Genome Research 21:2167-2180.
Our work uses functional genomics DNase-seq, ChIP-seq, RNA-seq, and chromatin state data to computationally identify combinations of transcription factor binding sites which operate to define the activity of cell-type specific enhancers. We are currently focused on:
- improving SVM methodology by including more general sequence features and constraints
- predicting the impact of SNPs on enhancer activity (delta-SVM) and GWAS association for specific diseases
- experimentally assessing the predicted impact of regulatory element mutation in mammalian cells
- systematically determining regulatory element logic from ENCODE human and mouse data
- using this sequence based regulatory code to assess common modes of regulatory element evolution and variation
We are located in the McKusick-Nathans Institute for Genetic Medicine, and the Department of Biomedical Engineering, which has long been a leader in the development of rigorous quantitative modeling of biological systems, and is a natural home for graduate studies in Bioinformatics and Computational Biology at Johns Hopkins, including research in Genomics, Systems Biology, Machine Learning, and Network Modeling.
Lab Members
Publications
Postdoctoral Positions Available
About Computational Biology in JHU Biomedical Engineering:
The Department of Biomedical Engineering has long been a leader in the development of rigorous quantitative modeling of biological systems, and is a natural home for graduate studies in Bioinformatics and Computational Biology at Johns Hopkins. Students with backgrounds in Physics, Mathematics, Computer Science and Engineering are encouraged to apply. Opportunities for research include: Computational Medicine, Genomics, Systems Biology, Machine Learning, and Network Modeling. Graduate students in Johns Hopkins' Biomedical Engineering programs can select research advisors from throughout Johns Hopkins' Medical Institutions, Whiting School of Engineering, and Krieger School of Arts and Sciences.